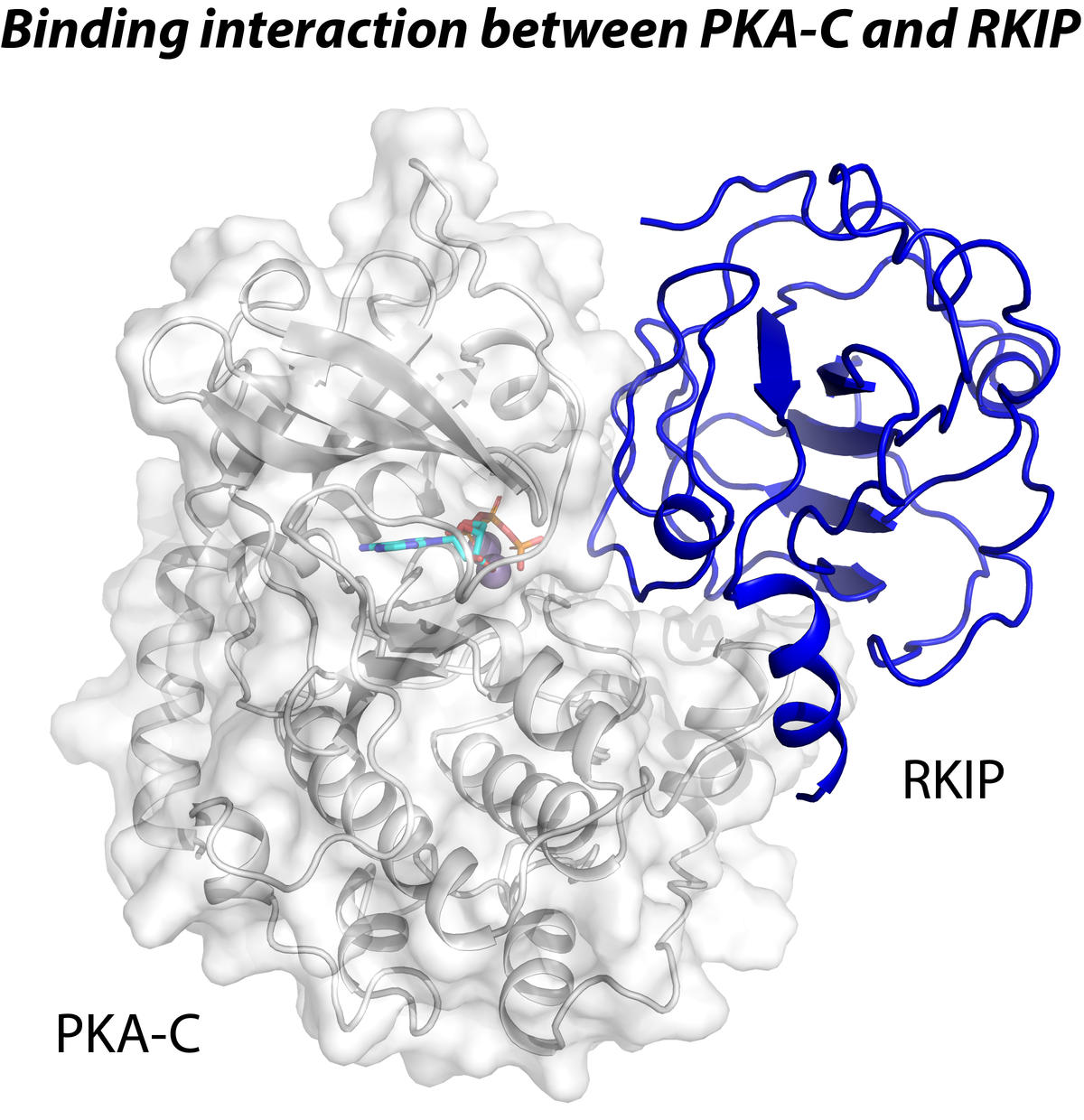
Overview
Our research focuses on two critical aspects of cell regulation: cAMP-mediated cell signaling and calcium transport. Soluble and integral membrane protein complexes orchestrate these events. We utilize a multidisciplinary approach combining solution and solid-state NMR spectroscopy with other biophysical methods to study these protein complexes' structure, dynamics, and interactions. Our goal is to understand how these protein complexes mediate allosteric signal transduction in cells and how mutations of these proteins are linked to diseases.